import dascore as dc
from dascore import print
pa1 = dc.get_example_patch("random_das")
pa2 = dc.get_example_patch("example_event_1")Patch
A Patch manages an array and its associated coordinate labels and metadata.
The Patch design was inspired by Xarray’s DataArray
Patch creation
Patches can be created in several different ways.
Load an example patch
DASCore includes several example datasets. They are mostly used for simple demonstrations and testing.
See get_example_patch for supported patches.
Load a file
A single file can be loaded like this:
Code
# This code block is only here to create a usable path for the next cell.
import dascore as dc
# Import fetch to read DASCore example files.
from dascore.utils.downloader import fetch
path = fetch("terra15_das_1_trimmed.hdf5")
# To read DAS data stored locally on your machine, simply replace the above line with:
# path = "/path/to/data/directory/data.EXT"import dascore as dc
# path should point to your file, e.g.
# path = mydata.hdf5
pa = dc.spool(path)[0]Spools are covered in more detail in the next section.
Manually create a patch
Patches can be created from:
- A data array
- Coordinates for labeling each axis
- Attributes (optional)
import numpy as np
import dascore as dc
from dascore.utils.time import to_timedelta64
# Create the patch data
array = np.random.random(size=(300, 2_000))
# Create attributes, or metadata
attrs = dict(
category="DAS",
id="test_data1",
data_units="um/(m * s)"
)
# Create coordinates, labels for each axis in the array.
time_start = dc.to_datetime64("2017-09-18")
time_step = to_timedelta64(1 / 250)
time = time_start + np.arange(array.shape[1]) * time_step
distance_start = 0
distance_step = 1
distance = distance_start + np.arange(array.shape[0]) * distance_step
coords = dict(time=time, distance=distance)
# Define dimensions (first label corresponds to data axis 0)
dims = ('distance', 'time')
pa = dc.Patch(data=array, coords=coords, attrs=attrs, dims=dims)Patch anatomy
Data
The data is simply an n-dimensional array which is accessed with the data attribute.
import dascore as dc
patch = dc.get_example_patch()
print(f"Data shape is {patch.data.shape}")
print(f"Data contents are\n{patch.data}")Data shape is (300, 2000)
Data contents are [[0.77770241 0.23754122 0.82427853 ... 0.36950848 0.07650396 0.23197621] [0.49689594 0.44224037 0.70329426 ... 0.12617754 0.11760625 0.78003741] [0.20681917 0.19516906 0.17434521 ... 0.84933595 0.36479426 0.80740811] ... [0.61877586 0.1053084 0.66896335 ... 0.621027 0.43559346 0.49975826] [0.75717115 0.25935121 0.09051709 ... 0.36099578 0.9365496 0.10351814] [0.15780837 0.29487104 0.58475197 ... 0.22898748 0.23950251 0.49439913]]
The data arrays should be read-only. This means you can’t modify them, but must first make a copy.
import numpy as np
patch.data[:10] = 12 # won't work
array = np.array(patch.data) # this makes a copy
array[:10] = 12 # then this worksCoords
DASCore implements a class called CoordManager which manages dimension names, coordinate labels, selecting, sorting, etc. CoordManager has several convenience methods for accessing contained information:
import dascore as dc
patch = dc.get_example_patch()
coords = patch.coords
# Get an array of time values
time_array = coords.get_array("time")
# Get the maximum distance value
distance_max = coords.max("distance")
# Get the time step (NaN if time isn't evenly sampled)
time_step = coords.step("time")For convenience, coordinates and their corresponding arrays can be accessed from the patch level as well.
import dascore as dc
patch = dc.get_example_patch()
# Get the coordinate object for distance
distance_coord = patch.get_coord("distance")
# Get the array of values corresponding to time
time_array = patch.get_array("time")Coords also have an expressive string representation:
print(coords)➤ Coordinates (distance: 300, time: 2000) *distance: CoordRange( min: 0 max: 299 step: 1 shape: (300,) dtype: int64 units: m ) *time: CoordRange( min: 2017-09-18 max: 2017-09-18T00:00:07.996 step: 0.004s shape: (2000,) dtype: datetime64[ns] units: s )
Patch dimensions may have an associated coordinate with the same name but this is not required.
Coordinates are often (but not always) associated with one or more dimensions. For example, coordinates “latitude” and “longitude” are often associated with dimension “distance”.
Most of the other CoordManager features are primarily used internally by DASCore, but you can read more about them in the Coordinate Tutorial.
Attrs
The metadata stored in Patch.attrs is a pydantic model which enforces a schema and provides validation. PatchAttrs.get_summary_df generates a table of the attribute descriptions:
| attribute | description |
|---|---|
| data_type | Describes the quantity being measured. |
| data_category | Describes the type of data. |
| data_units | The units of the data measurements |
| instrument_id | A unique id for the instrument which generated the data. |
| acquisition_id | A unique identifier linking this data to an experiment. |
| tag | A custom string field. |
| station | A station code. |
| network | A network code. |
| history | A list of processing performed on the patch. |
| dims | A tuple of comma-separated dimensions names. |
Specific data formats may also add attributes (e.g. “gauge_length”, “pulse_width”), but this depends on the parser.
String representation
DASCore Patches have a useful string representation:
import dascore as dc
patch = dc.get_example_patch()
print(patch)DASCore Patch ⚡ --------------- ➤ Coordinates (distance: 300, time: 2000) *distance: CoordRange( min: 0 max: 299 step: 1 shape: (300,) dtype: int64 units: m ) *time: CoordRange( min: 2017-09-18 max: 2017-09-18T00:00:07.996 step: 0.004s shape: (2000,) dtype: datetime64[ns] units: s ) ➤ Data (float64) [[0.778 0.238 0.824 ... 0.37 0.077 0.232] [0.497 0.442 0.703 ... 0.126 0.118 0.78 ] [0.207 0.195 0.174 ... 0.849 0.365 0.807] ... [0.619 0.105 0.669 ... 0.621 0.436 0.5 ] [0.757 0.259 0.091 ... 0.361 0.937 0.104] [0.158 0.295 0.585 ... 0.229 0.24 0.494]] ➤ Attributes tag: random category: DAS
Shortcuts
DASCore Patches offer a few shortcuts for quickly accessing commonly used information:
import dascore as dc
patch = dc.get_example_patch()
print(patch.seconds) # number of seconds in the patch
print(patch.channel_count) # number of channels in the patch8.0
300
These shortcuts only work for patches with dimensions “time” and “distance”, but they can help new users who may be unfamiliar with datetimes and coordinates.
The “name” of a patch (the filename DASCore would use if the patch were saved to disk) can be generated with Patch.get_patch_name. For a spool, use Spool.get_patch_names to get the names of its managed patches.
import dascore as dc
patch = dc.get_example_patch()
spool = dc.get_example_spool()
patch_name = patch.get_patch_name()
patch_names = spool.get_patch_names()Trim and Reshape
The following methods help trim, reshape, and manipulate coordinates.
Select
Patches are trimmed using the Patch.select method. Unlike Patch.order, select will not change the order of the affected dimensions, it will only remove elements. Most commonly, select takes the coordinate name and a tuple of (lower_limit, upper_limit) as the values. Either limit can be ... indicating an open interval.
import numpy as np
import dascore as dc
patch = dc.get_example_patch()
attrs = patch.attrs
# Select 1 sec after current start time to 1 sec before end time.
time = patch.get_coord("time")
one_sec = dc.to_timedelta64(1)
select_tuple = (time.min() + one_sec, time.max() - one_sec)
new = patch.select(time=select_tuple)
# Select only the first half of the distance channels.
distance_max = np.mean(patch.coords.get_array('distance'))
new = patch.select(distance=(..., distance_max))The relative keyword is used to trim coordinates based on the start (positive) and end (negative).
import dascore as dc
from dascore.units import ft
patch = dc.get_example_patch()
# We can make the example above simpler with relative selection
new = patch.select(time=(1, -1), relative=True)
# select 2 seconds from end to 1 second from end
new = patch.select(time=(-2, -1), relative=True)
# select last 100 ft of distance channels
new = patch.select(distance=(-100 * ft, ...), relative=True)The samples keyword tells select the meaning of the query is in samples rather than the units of the selected dimension. Unlike absolute selections, sample selections are always relative to the data contained in the patch. For example, 0 refers to the first sample along the dimension and -1 refers to the last.
import dascore as dc
patch = dc.get_example_patch()
# Trim patch to only include first 10 time rows (or columns).
new = patch.select(time=(..., 10), samples=True)
# Only include the last distance column or row.
new = patch.select(distance=-1, samples=True)Arrays can also be passed as values in which case they will be treated like sets, meaning only coordinate elements in the array will be selected.
import numpy as np
import dascore as dc
patch = dc.get_example_patch()
# Create an array of desired distances.
dist_to_select = np.array([10., 18., 12.])
sub_patch = patch.select(distance=dist_to_select)
# Test that select worked.
assert set(sub_patch.get_array('distance')) == set(dist_to_select)
# Samples also work
sub_patch = patch.select(distance=np.array([0, 12, 10, 9]), samples=True)
assert len(sub_patch.get_array('distance')) == 4Order
Order is similar to Patch.select, but will re-arrange data to the order specified by a value array. This may also cause parts of the patch to be duplicated.
import numpy as np
import dascore as dc
patch = dc.get_example_patch()
# Get a patch with a new distance ordering
dist_order1 = np.array([20., 10., 15.])
patch_dist1 = patch.order(distance=dist_order1)
assert np.all(dist_order1 == patch_dist1.get_array("distance"))
# Get a patch with duplicate entries for distance
dist_order2 = np.array([20., 20., 20.])
patch_dist2 = patch.order(distance=dist_order2)
assert np.all(dist_order2 == patch_dist2.get_array("distance"))New dimensions
Sometimes it can be useful to add new (empty) dimensions to a Patch. Patch.append_dims does this.
import dascore as dc
patch = dc.get_example_patch()
# Create a new patch with a single empty dimension called "nothing"
patch_dims = patch.append_dims("nothing")
# Create a new patch with a length two dimension called "money".
# The contents of the patch are repeated along the new dimension to fill
# the required length.
patch_extended_dims = patch.append_dims(money=2)
# Transpose can then be used to re-arrange the dims
patch_extended = patch_extended_dims.transpose("time", "money", "distance")
# And update coords to add coordinate values to the new dim
patch_extended_coord = patch_extended.update_coords(money=[10, 30])Although these examples are quite contrived, these functions are very useful for transforms which create high dimensional patches.
Processing
The patch has several methods which are intended to be chained together via a fluent interface, meaning each method returns a new Patch instance.
import dascore as dc
pa = dc.get_example_patch()
out = (
# Decimate to reduce data volume by 8 along time dimension
pa.decimate(time=8)
# Detrend along distance dimension
.detrend(dim='distance')
# Apply a low-pass 10 Hz butterworth filter along time dimension
.pass_filter(time=(..., 10))
)The processing methods are located in the dascore.proc module. The patch processing tutorial provides more information about processing routines.
Visualization
DASCore provides some visualization functions in the dascore.viz module or using the Patch.viz namespace. DASCore generally only implements simple, matplotlib based visualizations but other DASDAE packages will likely do more interesting visualizations.
import dascore as dc
patch = (
dc.get_example_patch('example_event_1')
.taper(time=0.05)
.pass_filter(time=(None, 300))
)
patch.viz.waterfall(show=True);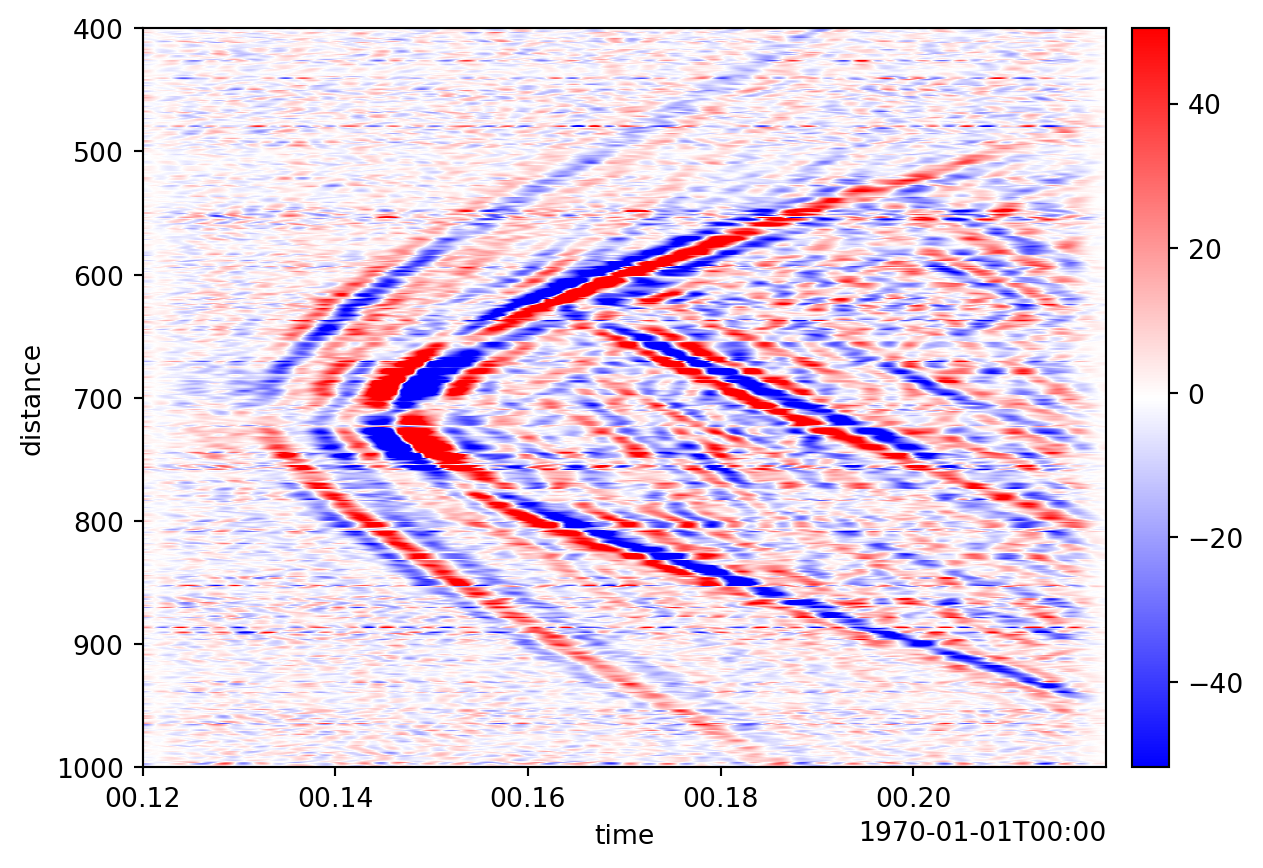
Modifying patches
Because patches should be treated as immutable objects, they can’t be modified with normal attribute assignment. However, DASCore provides several methods that return new patches with modifications.
Update
Patch.update uses the Patch instances as a template and returns a new Patch instances with one or more aspects modified.
import dascore as dc
pa = dc.get_example_patch()
# Create a copy of patch with new data but coords and attrs stay the same.
new_data_patch = pa.update(data=pa.data * 10)
# Completely replace the attributes.
new_data_patch = pa.update(attrs=dict(station="TMU"))Update attrs
Patch.update_attrs is for making changes to the attrs (metadata) while keeping the unaffected metadata (Patch.update would completely replace the old attrs).
import dascore as dc
pa = dc.get_example_patch()
# Update existing attribute 'network' and create new attr 'new_attr'
pa1 = pa.update_attrs(network='exp1', new_attr=42)Update coords
Patch.update_coords returns a new patch with the coordinates changed in some way. These changes can include: - Modifying (updating) existing coordinates - Adding new coordinates - Changing coordinate dimensional association
Modifying coordinates
Coordinates can be updated by specifying a new array which should take the place of the old one:
import dascore as dc
pa = dc.get_example_patch()
# Add one second to all values in the time array.
one_second = dc.to_timedelta64(1)
old_time = pa.coords.get_array('time')
new = pa.update_coords(time=old_time + one_second)Or by specifying new min, max, or step values for a coordinate.
import dascore as dc
pa = dc.get_example_patch()
# Change the starting time of the array.
new_time = pa.coords.min('time') + one_second
new = pa.update_coords(time_min=new_time)Adding coordinates
Commonly, additional coordinates, such as latitude/longitude, are attached to a particular dimension such as distance. It is also possible to include coordinates that are not associated with any dimensions.
import numpy as np
import dascore as dc
pa = dc.get_example_patch()
coords = pa.coords
dist = coords.get_array('distance')
time = coords.get_array('time')
# Add a single coordinate associated with distance dimension.
lat = np.arange(0, len(dist)) * .001 -109.857952
# Note the tuple form: (associated_dimension, value)
out_1 = pa.update_coords(latitude=('distance', lat))
# Add multiple coordinates associated with distance dimension.
lon = np.arange(0, len(dist)) *.001 + 41.544654
out_2 = pa.update_coords(
latitude=('distance', lat),
longitude=('distance', lon),
)
# Add coordinate associated with multiple dimensions.
quality = np.ones_like(pa.data)
out_3 = pa.update_coords(
quality=(pa.dims, quality)
)
# Add coordinate which isn't associated with a dimension.
no_dim_coord = pa.update_coords(non_dim=(None, np.arange(10)))Changing coordinate dimensional association
The dimensions each coordinate is associated with can be changed. For example, to remove a coordinate’s dimension association:
import dascore as dc
# Load a patch which has latitude and longitude coordinates.
patch = dc.get_example_patch("random_patch_with_lat_lon")
# Disassociate latitude from distance.
lat = patch.coords.get_array('latitude')
patch_detached_lat = patch.update_coords(latitude=(None, lat))Dropping coordinates
Non-dimensional coordinates can be dropped using Patch.drop_coords. Dimensional coordinates, however, cannot be dropped doing so would force the patch data to become degenerate.
import dascore as dc
# This patch has latitude and longitude coordinates
patch = dc.get_example_patch("random_patch_with_lat_lon")
# Drop latitude, this won't affect the data or other coordinates
patch_dropped_lat = patch.drop_coords("latitude")
print(patch_dropped_lat.coords)➤ Coordinates (distance: 300, time: 2000) *distance: CoordRange( min: 0 max: 299 step: 1 shape: (300,) dtype: int64 units: m ) *time: CoordRange( min: 2017-09-18 max: 2017-09-18T00:00:07.996 step: 0.004s shape: (2000,) dtype: datetime64[ns] units: s ) longitude ('distance',): CoordRange( min: 41.5 max: 41.8 step: 0.001 shape: (300,) dtype: float64 )
Coords in patch initialization
Any number of coordinates can also be assigned when the patch is initiated. For coordinates other than those of the patch dimensions, the associated dimensions must be specified. For example:
import dascore as dc
import numpy as np
# Create data for patch
rand = np.random.RandomState(13)
array = rand.random(size=(20, 100))
time1 = np.datetime64("2020-01-01")
# Create patch attrs
attrs = dict(dx=1, d_time=1 / 250.0, category="DAS", id="test_data1")
time_deltas = dc.to_timedelta64(np.arange(array.shape[1]) * attrs["d_time"])
# Create coordinate data
distance = np.arange(array.shape[0]) * attrs["dx"]
time = time1 + time_deltas
quality = np.ones_like(array)
latitude = np.arange(array.shape[0]) * .001 - 111.00
# Create coord dict
coords = dict(
distance=distance,
time=time,
latitude=("distance", latitude), # Note distance is attached dimension
quality=(("distance", "time"), quality), # Two attached dimensions here
)
# Define dimensions of array and init Patch
dims = ("distance", "time")
out = dc.Patch(data=array, coords=coords, attrs=attrs, dims=dims)Units
As mentioned in the units section of the concept page, DASCore provides first-class support for units.
Patch units
There are two methods for configuring the units associated with a Patch.
Patch.set_units sets the units on a patch or its coordinates. Old units are simply overwritten without performing any conversions. The first argument sets the data units and the keywords set the coordinate units.
Patch.convert_units converts data or coordinates units by appropriately transforming the data or coordinates arrays. If no units exist they will simply be set.
import dascore as dc
patch = dc.get_example_patch()
# Set data units and distance units; don't do any conversions
patch_set_units = patch.set_units("m/s", distance="ft")
# Convert data units and distance units; will modify data/coords
# to correctly do the conversion.
patch_conv_units = patch_set_units.convert_units("ft/s", distance='m')The data or coordinate units attributes are Pint Quantity, but they can be converted to strings with get_quantity_str.
import dascore as dc
from dascore.units import get_quantity_str
patch = dc.get_example_patch().set_units("m/s")
print(type(patch.attrs.data_units))
print(get_quantity_str(patch.attrs.data_units))<class 'pint.Quantity'>
m / s
Units in processing functions
import dascore as dc
from dascore.units import m, ft
pa = dc.get_example_patch()
# Sub-select a patch to only include distance from 10ft to 10m.
sub_selected = pa.select(distance=(10*ft, 10*m))
# Filter patch for spatial wavelengths from 10m to 100m.
dist_filtered = pa.pass_filter(distance=(10*m, 100*m))See the documentation on Patch.select and Patch.pass_filter for more details.
Patch operations
Patches implement many numpy-like functions which are applied directly to a patch using built-in python operators.
In the case of scalars and numpy arrays, the operations are broadcast over the patch data. In the case of two patches, compatibility between patches are first checked, the intersection of the coords and attrs are calculated, then the operator is applied to both patches’ data. Here are a few examples:
See merge_compatible_coords_attrs for more details on how attributes and coordinates are handled when performing operations on two patches.
Patch operations with scalars
import numpy as np
import dascore as dc
patch = dc.get_example_patch()
out1 = patch / 10
assert np.allclose(patch.data / 10, out1.data)
out2 = patch ** 2.3
assert np.allclose(patch.data ** 2.3, out2.data)
out3 = patch - 3
assert np.allclose(patch.data - 3, out3.data)Units are also fully supported.
import dascore as dc
from dascore.units import m, s
patch = dc.get_example_patch().set_units("m/s")
# Multiplying patches by a quantity with units updates the data_units.
new = patch * 10 * m/s
print(f"units before operation {patch.attrs.data_units}")
print(f"units after operation {new.attrs.data_units}")units before operation 1.0 m / s
units after operation 1.0 m ** 2 / s ** 2
Patch operations with numpy arrays
Patch implements the numpy array protocol, meaning you can use numpy functions on patches
import numpy as np
import dascore as dc
patch = dc.get_example_patch()
ones = np.ones(patch.shape)
out1 = patch + ones
assert np.allclose(patch.data + ones, out1.data)Units also work with numpy arrays.
import numpy as np
import dascore as dc
from dascore.units import furlongs
patch = dc.get_example_patch()
ones = np.ones(patch.shape) * furlongs
out1 = patch * ones
print(f"units before operation {patch.attrs.data_units}")
print(f"units after operation {out1.attrs.data_units}")units before operation None
units after operation 1 fur
Patch operations with other patches
Identically shaped patches
When patches are shaped the same, the operations can simply be applied on the data arrays, then coords and attrs merged.
import numpy as np
import dascore as dc
from dascore.units import furlongs
patch = dc.get_example_patch()
# Adding two patches together simply adds their data their
# and checks/merges coords and attrs.
out = patch + patch
assert np.allclose(patch.data * 2, out.data)Broadcastable patches
If the arrays are not shaped the same, but they can be broadcasted to compatible shapes, these operations work as well.
import numpy as np
import dascore as dc
from dascore.units import furlongs
patch = dc.get_example_patch()
sub_patch = patch.select(distance=1, samples=True)
print(sub_patch.shape)
# The sub patch is broadcasted over the patch along appropriate dimension.
sum_patch = patch + sub_patch
print(sum_patch.shape)(1, 2000)
(1, 2000)
Numpy Functions
DASCore uses apply_array_ufunc and array_function for applying numpy functions to Patch instances.
For NumPy’s ufuncs DASCore implements Patch universal functions. You can use numpy ufuncs directly on patches or create patch-specific ufuncs with dimension support.
import numpy as np
import dascore as dc
patch = dc.get_example_patch()
# Use numpy ufuncs directly on patches
abs_patch = np.abs(patch)
sin_patch = np.sin(patch)
# Add patches together using numpy ufuncs
sum_patch = np.add(patch, patch)Some of the Patch methods are ufuncs which means they support accumulation and reduction along specified dimensions.
from dascore.utils.array import PatchUFunc
import numpy as np
import dascore as dc
patch = dc.get_example_patch()
added_patch = patch.add(patch)
patch_cum_sum = patch.add.accumulate("time")
patch_reduced = patch.add.reduce("distance")PatchUFunc.reduce/accumulate accept dim (mapped internally to axis), while numpy ufuncs accept axis.For more advanced usage, you can create patch ufuncs.
from dascore.utils.array import PatchUFunc
import numpy as np
import dascore as dc
# Create a patch ufunc wrapper
add_ufunc = PatchUFunc(np.add)
patch = dc.get_example_patch()
# Use accumulate to compute cumulative sum along time dimension
cumsum_patch = add_ufunc.accumulate(patch, dim="time")
# Use reduce to sum all values along distance dimension
sum_patch = add_ufunc.reduce(patch, dim="distance")
# Bind to patch for cleaner syntax
bound_add = add_ufunc.__get__(patch, type(patch))
result = bound_add.reduce(dim="time") # No need to pass patchThe patch ufunc system automatically handles dimension-to-axis conversion and preserves patch coordinates appropriately.
